The evolution of silicon transporters in diatoms. Journal of Phycology (html)(pdf)
study co-authors: Julie Koester (UNC Wilmington), Sara Bender (Gordon and Betty Moore Foundation), E. Virginia Armbrust (UW)
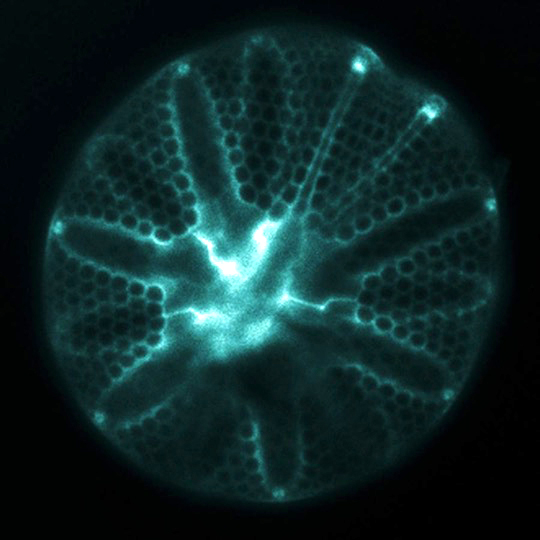
The ornately patterned silica cell walls (“frustules”) formed by diatoms attract attention for their beauty, but also give diatoms a distinct and globally important role in the oceans. These glass cell walls are created from dissolved silicon (silicic acid) that is actively taken up from seawater by the cells. Diatoms can not grow and divide without first forming this silica frustule, so their ability to take up silicic acid from seawater is a controlling factor on the magnitude of primary production that fuels ocean food webs and also influences global-scale biogeochemistry.
When diatoms evolved, the concentration of silicic acid in the ocean was likely much higher than today, and was drastically reduced as diatoms proliferated and diversified. The molecular machinery used to take up silicic acid may therefor include both relicts that evolved in an ancient ocean environment, and more recent adaptations that enable diatoms to thrive in the modern low-Si ocean.
We recently published a study examining the evolution of the proteins that enable diatoms to activley transport silicic acid into the cell: silicon transporters or “SITs”. This was made possible due to the recently sequenced, publicly available, community-produced transcriptomes of ~750 marine microeukaryotes. The diversity of organisms within this dataset enabled us to identify more diverse gene sequences across a broader range of organisms than previously possible.
First, we made a phylogenetic tree to model the evolutionary relationships among the gene sequences. In my opinion, this was the most challenging and critical part of the study because we wanted to use the most conservative methods to produce the most accurate tree, without sacrificing the inclusion of incredibly diverse sequences in the large transcriptome database. We eventually figured it out, but you’ll have to read the methods in the paper if you’re curious.
(Phylogeny of 473 SIT genes identified in this study)
![]() This tree was much too big to analyze manually, which was a great motivation to learn python. (I used the ETE2 tools in python to study this tree.) Every “branch” on the tree is a gene sequence encoded by an organism, and branches that are connected are more closely related to one another. The nodes where branches connect represent a common ancestral gene that would have existed in the past.
This tree was much too big to analyze manually, which was a great motivation to learn python. (I used the ETE2 tools in python to study this tree.) Every “branch” on the tree is a gene sequence encoded by an organism, and branches that are connected are more closely related to one another. The nodes where branches connect represent a common ancestral gene that would have existed in the past.
One exciting discovery that emerged from this tree was the presence of “SITs” in other marine protists, not diatoms. These sequences clustered together, apart from the diatoms, but shared a common ancestral sequence with diatoms. This suggested that silicon plays a role in the cell biology of other marine protists (such as in a recent study of coccolithophores), and also that the earliest diatoms likely inherited SITs from an ancestral protist when they emerged in a high-Si ocean. Actively taking up Si is clearly an important function for the cell, even in high Si conditions, and we identified decendants of these earliest SIT genes in all major diatom lineages (black branches).
A second interesting discovery was that diatom SIT genes continued to diverge and diversify as diatom species evolved. When a gene duplicates in the genome, the duplicate sequence can change and has the potential to take on new functions. We identified 5 distinct types of SIT genes (branches on the tree are color-coded by type), and each species encodes its own repertoire of SIT gene types. The diatoms that evolved last (pennates) encode the most diverse combination of SIT sequence types. The diatoms that evolved first (centrics) encode the
least diverse combination of SIT sequence types.
This is very exciting because of the potential ecological ramifications: different diatom lineages may have different abilities to take up silicic acid, and they may have distinct responses to changes in its availability. Encoding different SIT gene repertoires may be a molecular basis for niche-partitioning among co-exisiting species that need to utilize the same resource (Si).
The ecological implications of the evolutionary history of SITs were intriguing, and we wanted to know what type of SIT genes are used by natural communities of diatoms, and whether it changes depending on the species present and the environmental condition. To look for evidence that different SIT types are used in different conditions, we studied genetic samples from 4 locations off the coast of Washington and British Columbia (collected during the 2012 GeoMICS cruise). These samples represent all the genes transcribed (i.e.
“expressed”) by natural mixed plankton communities at the time of sampling (i.e. metatranscriptomes). Because we had such an extensive phylogenetic tree as a reference, we were able to identify environmental sequences by their SIT sequence type and also associate those sequences with organisms that have closely related sequences.
Pie charts show SIT sequence types (different colors) expressed by different diatom lineages at two different locations.
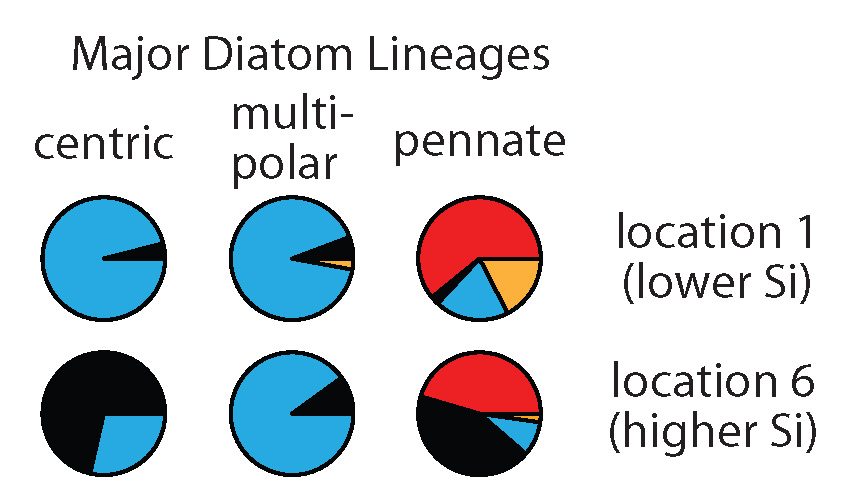
We found large differences in the type of SITs most expressed by different diatom lineages (compare pies horizontally). We also saw changes in the type of SITs expressed within these lineages between locations (compare pies vertically). Interestingly, at the location with the highest silicic acid, we detected relatively more of the earliest SIT type most closely related to the genes encoded by the first diatoms that lived in a high Si ocean (black pie pieces). In this study, we don’t have the data to definitively explain why diatoms changed the types of SITs expressed between environments, but the data do suggest that these measurements could be useful in future studies that try to resolve the responses and physiology of natural populations.
This project was a collaboration with the colleagues I worked with during my PhD at UW. We came up with the idea for this project just as we all moved to different parts of the country, but our enthusiasm for the topic is what kept us working on it over the next four years, often in our free time. Thank you to my “SIT-sters”!


